
Nitin Uttam Kamble
PostDoc MECEA Winner 2024
From Seed to Starch: Unravelling Mysteries for Sustainable Agriculture and Industry.
- PostDoc MECEA Winner 2024
- | Part of MECEA
Growing up in a farming family in a small village in India, I witnessed the challenges farmers face in achieving successful crop yields and preserving their harvests, which motivated me to find sustainable solutions for long-term grain storage and utilization. This led me to pursue a Ph.D. at National Institute of Plant Genome Research (NIPGR) in New Delhi, India where I unravelled the important role of protein modifications in enhancing seed germination vigor and viability in rice [1-3]. This pivotal work aimed to understand the underlying mechanisms influencing seed quality, a critical factor in improving crop productivity and ensuring effective post-harvest storage.
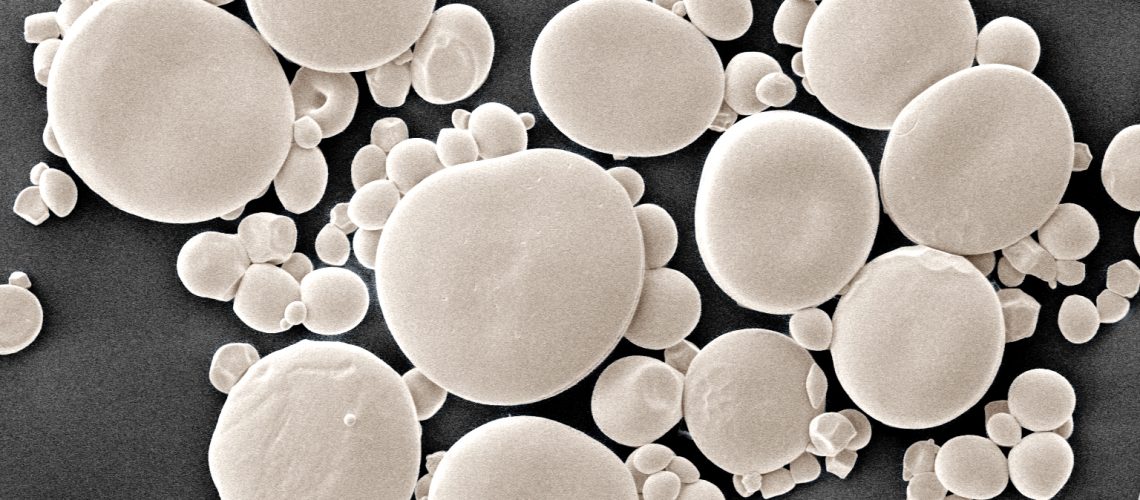
During my time at NIPGR, I had the opportunity to join David Seung’s lab at the prestigious John Innes Centre in the United Kingdom as a visiting scientist in 2021. During this time, I discovered the captivating world of starch and the important role it plays in grain quality. Starch consists of glucose polymers amylopectin and amylose assembled into tiny semi-crystalline granules. Wheat and other Triticeae species exhibit two distinct granule types: large A-type granules and smaller B-type granules. The work of Erica Hawkins and colleagues had shown that STARCH SYNTHASE 4 (SS4) and B-GRANULE CONTENT1 (BGC1) are required for proper A-type granule initiation in wheat [4], and our collaborator Kay Trafford’s lab at NIAB had demonstrated that BGC1 also plays a crucial role in B-type granule initiation[5]. I was therefore motivated to investigate the mechanism by which BGC1 acts in B-type granule initiation.
Drawing on my previous expertise in protein biochemistry, I decided to look for protein interaction partners for BGC1. Using a pulldown followed by Mass spectrometry, I discovered that BGC1 interacts with the glucan phosphorylase (PHS1) and a large number of other proteins. I focused on the interaction between PHS1 and BGC1, given prior knowledge that PHS1 can elongate glucan substrates. Utilizing the wheat TILLING collection, I isolated durum wheat mutants deficient in PHS1, which exhibited normal A-type granules but fewer and larger B-type granule [6]. Surprisingly, starch content, grain size, and grain yield per plant were not affected by the PHS1 mutation. My discovery is significant as the role of PHS1 has been debated since the 1940s, and now I can assign the first specific function to PHS1 in B-type granule initiation. I had demonstrated that A and B-type granule initiation occur via different enzymes.
I realized that understanding the distinct biochemical mechanisms that initiate A- and B-type granules in wheat is crucial for creating starch variations tailored for diverse food and industrial applications. The coexistence of these granule types poses a significant challenge for the starch manufacturing industry, as many smaller B-type granules are lost and wasted during milling. The ratio of A- and B-type granules also impacts the quality of wheat-based foods, such as bread and pasta. Moreover, an excess of B-type starch granules in barley can lead to a hazy or cloudy appearance in beer due to their resistance to digestion and filtration during brewing. Now, I can use my discovery to manipulate the ratios between A- and B- granules in wheat and other Triticeae crops.
I am currently investigating whether the other BGC1 interaction partners identified in our study are involved in B-type granule initiation and how the actions of these proteins are coordinated. Additionally, I am also exploring how having fewer and larger B-type granules affect cooking quality and other starch properties. We are collaborating with a major European seed company and an international malting company to bring my discoveries to application. I hope my work will be used to create useful starch variations, to achieve tailor-made starches for diverse food and industrial applications.
References:
[1] Petla et al. (2016). New Phytologist. 211:627-645.
[2] Kamble and Majee (2022). Development. 149 (11).
[3] Kamble et al. (2022) Environmental and Experimental botany. 202:105027.
[4] Hawkins et al. (2021) New Phytologist. 230, 2371-2386.
[5] Chia et al. (2020) Journal of Experimental Botany. 71, 105-115.
[6] Kamble et al. (2023). The Plant Cell. 1:20.
