
Anna Backhaus
PhD MECEA Winner 2022
Increasing future wheat yields, one spikelet at a time
- PhD MECEA Winner 2022
- | Part of MECEA
When I was 13 years old, I spoke to the 2008 UN climate change conference. There I learned about the intertwined relationship of climate change, agriculture, and global food security. I concluded that more efficient and resilient plants could be the key to safeguard the planets biodiversity and global food production. And I decided that I would like to spend my career helping this cause. I set my path travelling through maize fields at Purdue, wheat fields at JIC1, and surrounded by rusts in Morocco2. Ultimately, I focused my journey to the wonders of inflorescence development in the grasses. Under the right environmental conditions, the apical meristem of grasses switches from vegetative to floral development and starts to initiate spikelet primordia. Each spikelet will develop into multiple florets, however the number of initiated spikes, spikelets and florets can be constantly adapted to the environmental conditions throughout the plants’ development. Therefore, to understand what determines yield, we need to understand the genetic network in control of its development.
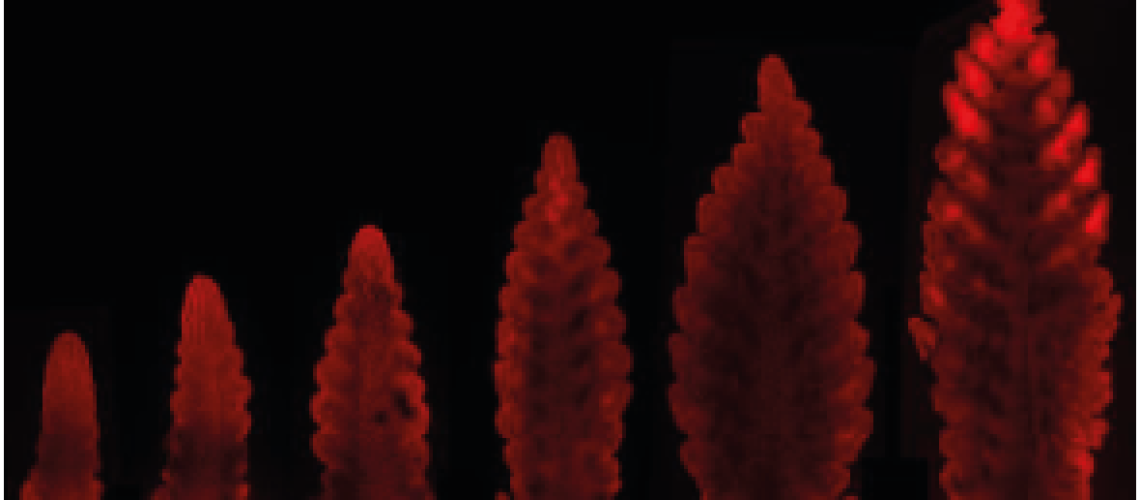
In 2018 I gained a place in the JIC rotation PhD program, which gives students the opportunity to undertake three brief projects before choosing their main PhD. During my rotations I was happily working on a range of topics, for example the role of NAC genes in leaf senescence3,4. But in the end, my obsession with the development of grass inflorescences was too strong. Much research has focused on increasing overall spikelet fertility. However, not all spikelets across a spike are equal: Central spikelets produce more and larger grains than their apical and basal counterparts. Often, the most basal spikelet is so small that it fails to produce any grain. The apical gradient can be explained as the spikelets are developed last and are furthest away from the peduncle. But why the basal spikelets are smaller, despite being initiated first (early stages in Fig 1), remained unanswered. This unexplained variation in spikelet productivity deeply fascinated me.
To tackle this mystery, I combine the strengths of three of my rotations. I realised quickly that basal spikelets fall back in early in development when the spike is only a few microns long. To study the transcriptional differences between the spikelets at this point the available RNA extraction methods were not sufficient to sequence these tiny tissues. Through collaborating with the Earlham institute, we were able to employ a novel, low-input RNA-seq method and were thus able to discover that thousands of genes are differentially expressed across a single spike5. These gradients were overlooked in previous studies due to the bulking of multiple, complete spikes for sequencing.
It was again the collaborative nature of my PhD project that helped us to identify genes affecting specifically the development of basal spikelets. Firstly, we modelled the molecular interaction of two of our candidate genes and found this sufficient to produce a lanceolate shaped wheat spike in silico (you can download and play with the model here!5). Secondly, one of the genes in our model had recently been characterised by a post-doc in our group and I thus had plenty of genetic material (NILs and transgenics) to test the gene’s effect on basal spikelets6. Excitingly, overexpression of the gene significantly decreased basal spikelet survival! Today I am entering new collaborative projects using live microscopy and specialised image processing algorithms to further investigate how the switch from one organ (leaves) to the next (spikelets) is orchestrated. But I am also always on the lookout for opportunities to share my fascination of plant development with others. I have created a series of comic books about plant biology for schools (funded by ASPB), written a blog about the history of studying plants and taught plant biology at primary school and university level. Even more I enjoy sharing my research with others through public speaking at conferences, in local schools or at outreach events. Studying plants might not be rocket science, but rocket scientist will undoubtedly rely on plant scientists to feed them in the future, either in this world or beyond.
References
1: “An analysis of Pseudomonas genomic diversity in take‐all infected wheat fields reveals the lasting impact of wheat cultivars on the soil microbiota.” TH Mauchline, D Chedom‐Fotso, G Chandra, T Samuels, N Greenaway, AE Backhaus, V McMillan, G Canning, SJ Powers, KE Hammond‐Kosack, PR Hirsch, IM Clark, Z Mehrabi, J Roworth, J Burnell, Jacob G Malone Environmental microbiology, (2015): 4764-4778 https://doi.org/10.1111/1462-2920.13038
2: “Genome-wide association study for adult plant resistance to yellow rust in spring bread wheat (Triticum aestivum L.).” S El Hanafi, AE Backhaus, N Bendaou, M Sanchez-Garcia, A Al-Abdallat, W Tadesse, Euphytica, 2021, 217(5), 1-14, https://doi.org/10.1007/s10681-021-02803-1
3: “A heat-shock inducible system for flexible gene expression in cereals.” SA Harrington, AE Backhaus, S Fox, C Roger, P Borrill, C Uauy, A Richardson, Plant Methods, 2020, https://doi.org/10.1186/s13007-020- 00677-3
4: “The Wheat GENIE3 Network Provides Biologically-Relevant Information in Polyploid Wheat” SA Harrington, AE Backhaus, A Singh, K Hassani-Pak, C Uauy, G3 Genes|Genomes|Genetics, 2020, https://doi.org/10.1534/g3.120.401436
5: “High expression of MADS-box VRT2 increases the number of rudimentary basal spikelets.” AE Backhaus, A Lister, M Tomkins, NM Adamski, J Simmonds, IC Macaulay, RJ Morris, W Haerty, C Uauy, 2022. Plant Physiology, 2022, https://doi.org/10.1093/plphys/kiac156
6: “Ectopic expression of Triticum polonicum VRT-A2 underlies elongated glumes and grains in hexaploid wheat in a dosage-dependent manner” NM Adamski, J Simmonds, JF Brinton, AE Backhaus, Y Chen, M Smedley, S Hayta, T Florio, P Crane, P Scott, A Pieri, O Hall, E Barclay, M Clayton, JH Doonan, C Nibau, C Uauy, The Plant Cell, 2021, https://doi.org/10.1093/plcell/koab119
