
Katie Long
PhD MECEA Winner 2025
Mapping the wheat spike – a journey through space and time
- PhD MECEA Winner 2025
- | Part of MECEA
I’ve always been fascinated by development—how seemingly simple structures become complex. While I enjoyed learning about zebrafish and Drosophila in university, plants stood out. Unable to move around their environment, they rely on remarkable adaptability. Even at the cellular level, plant cells cannot migrate within tissues and must coordinate with one another to grow and pattern organs. While some see plants as quite ‘passive’, I have always found them to be remarkably clever- making studying of their development all the more intriguing.
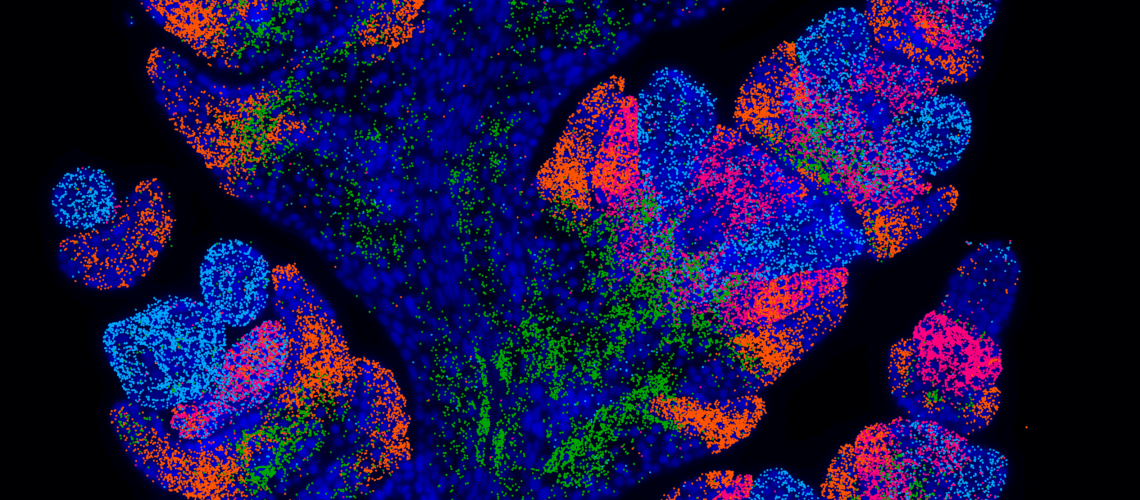
A crucial part of studying development is determining when and where genes are transcribed. I’ve been fortunate to study biology at a time of technological advancement that can resolve the transcriptome down to individual cells. First came single-cell RNA-seq, which captures the cell’s transcriptome at the expense of spatial context, as it requires tissue dissociation. Spatial transcriptomics has since revolutionized the field by preserving tissue architecture while mapping gene expression, allowing researchers to determine not only which genes a cell transcribes but also its precise location within a tissue. As spatial techniques became accessible to plant researchers, we saw the wheat inflorescence (or spike) as an ideal system for its application. The spike begins as a series of sequentially initiated meristems, which will differentiate into the spikelets. However, each of these spikelets will be slightly staggered in their progress- at any given point in development, there will be a range of spikelets at varying developmental stages. Spatial transcriptomics provided a unique opportunity to ‘map’ gene expression across the spike- retaining the context of each cell within this complex structure.
To accomplish this, we collaborated with the Earlham Institute, who were establishing a platform for spatial transcriptomics using VizGen MERSCOPE technology. This utilises an imaging-based technique called MERFISH that detects the precise location of individual transcripts and multiplexes this process to allow for hundreds of genes to be assayed in one experiment. This collaboration connected us with Ashleigh Lister, whose dedication enabled us to make large breakthroughs in troubleshooting and adapting MERFISH for notoriously challenging plant tissues. Preparing for this project was no minor feat- I would dissect up to a hundred plants a day- and meticulously line up tiny wheat spikes in a mould that we froze into blocks. With these, I prepared 10-micron thin sections, ensuring precise alignment to capture each inflorescence at the correct angle, and carefully transferred them onto delicate glass slides. It was nerve-wracking work to say the least! After this, Ashleigh’s team at Earlham would take over, prepping these samples for imaging on the VizGen MERSCOPE instrument.
Despite the challenges that tricky plant tissues threw our way, we successfully mapped the expression of 200 genes across wheat spikes to cellular resolution. I hope this work provides researchers with valuable gene expression data while also promoting the adoption of spatial techniques through our optimized methods. To ensure easy interaction with this dataset, we launched the platform wheat-spatial.com with the help of Rob Ellis at JIC. Please have a look for your favourite gene or just explore the dataset! Personally, one of my favourite parts of this project was the data analysis. For all those interested in how we can extract insights from microscopy images, we’ve made all raw data and scripts available. Finally, to dive deep into the developmental questions we addressed with this dataset, please see our pre-print.
